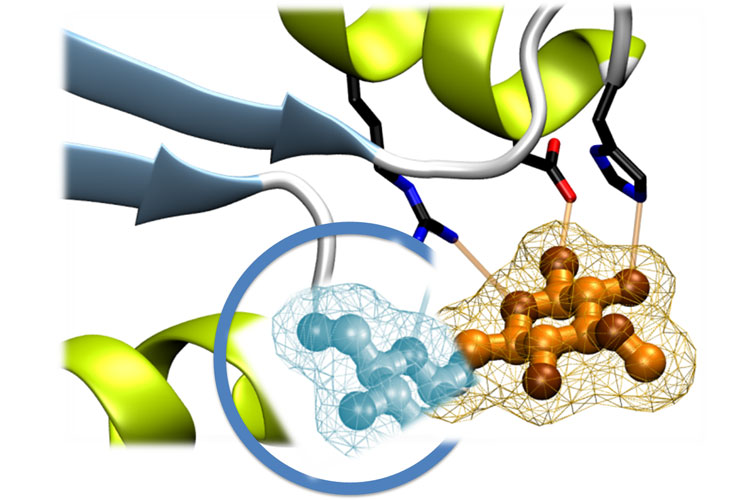
Enzymes catalyse a large number of biochemical reactions with remarkable efficiency and specificity thanks to their ability to dynamically rearrange the catalytic environment of reactions on natural substrates. However, for unconventional chemical transformations, enzyme-substrate interactions are not optimized and there is great room for improvement in catalytic efficiency. There is growing demand for efficient, selective, and stable enzymes to be applied in sustainable chemical transformation processes. This demand has driven great advances in computational enzymatic engineering, which has turned it into a scientific revolution, facilitating the development of new biocatalysts, for example to degrade plastics or for the synthesis of antibiotics.
At the IQS Biochemistry Laboratory, a multidisciplinary laboratory led by Dr Antoni Planas under the Biological and Biotechnological Chemistry Group (GQBB), the Bioinformatics and Molecular Modelling unit, coordinated by Dr Xevi Biarnés, is working on the development and application of computational biology and simulation methods based on artificial intelligence systems, data science, and high-performance calculations. One of the tools that this unit has created is the BINDSCAN protocol for enzyme design and optimization. This tool makes it possible to optimize the sequence of a starting biocatalyst in order to improve its performance and reaction efficiency on substrates that are not necessarily natural.
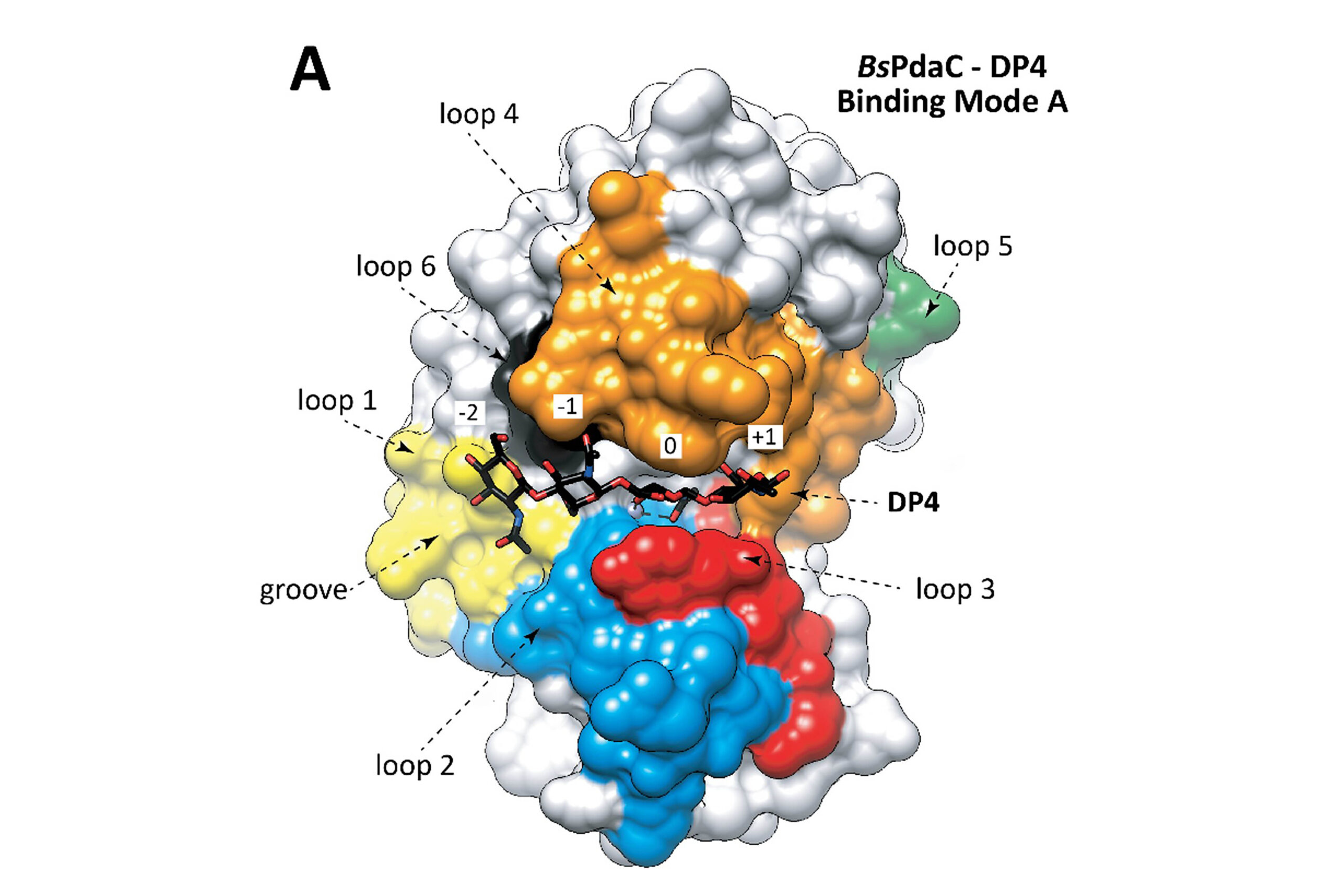
Within the GQBB group, one of their research aims is focused on glycosyl hydrolases (GH), enzymes that catalyse the cleavage of the glycosidic bond in glycans and glyconjugates. The reengineering of the enzymatic function of GH has enabled these researchers to produce high value-added carbohydrates such as prebiotics for formula, polysaccharides for nanomaterials, or plant stimulants, among others.
Within this broad context, and with the aim of expanding the prediction capabilities of the BINDSCAN tool, Dr Aitor Vega Sánchez conducted his doctoral thesis under the title BINDSCAN: a computational protocol for the engineering of glycosyl hydrolases, which was supervised by Dr Xevi Biarnés Fontal and Dr Antoni Planas Sauter within the IQS Department of Bioengineering.
New computational protocol based on massive mutational analysis of enzymes
Dr Vega’s thesis has established, for the first time, a robust computational protocol that makes it possible to quantify the electrostatic contribution of enzymes to their catalytic efficiency. Through massive mutational analysis of protein sequences, this approach identifies key positions that can be modified to improve functional properties, such as recognizing new ligands or catalytic performance, thus expanding the capabilities of the BINDSCAN tool for designing biocatalysts for chemical transformations.
As part of his research, Dr Vega developed an advanced version of the BINDSCAN tool that integrates innovative metrics based on binding affinity and electrostatic potential along with a graphical interface that facilitates the interpretation of the results.
This thesis represents a significant advance in integration between computational design and experimental validation and expands the potential of enzymatic engineering in industrial and biotechnological applications.
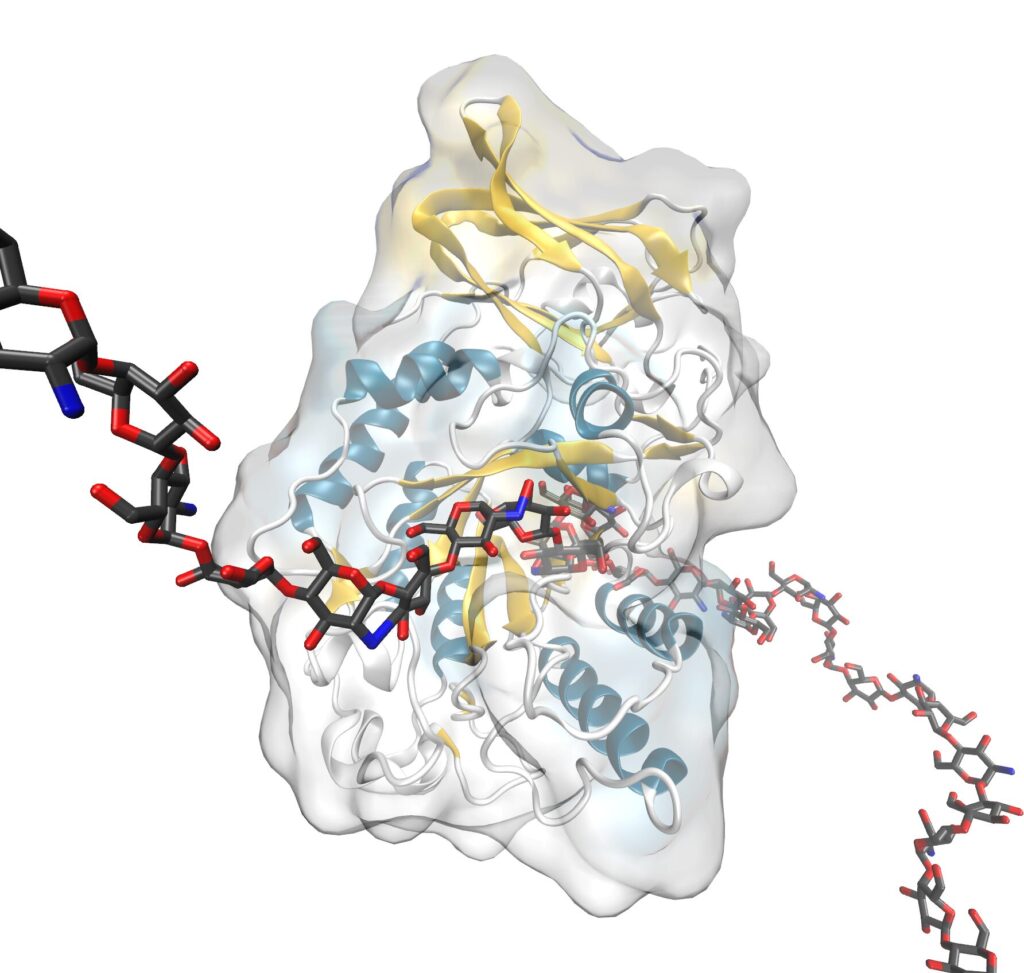
Within the GQBB group itself, this new BINDSCAN protocol has been applied to optimize the enzymatic synthesis of human breast milk oligosaccharides (HMOs), based on the redesign of the bacterial enzyme lacto-N-biosidase from Bifidobacterium bifidum, for the synthesis of lacto-N-tetraose (LNT), the main class I HMO.
In addition, BINDSCAN has also been applied to the redesign of other synthetic enzymes, in collaboration with researchers from the Toulouse Institute of Biotechnology (TBI), facilitating the production of high value-added plant origin oligosaccharides.
Related publications
AitorVega, Antoni Planas, Xevi Biarnés, Electrostatic potential as a reactivity scoring function in computer-assisted enzyme engineering, 2025, The FEBS Journal
Aitor Vega, Antoni Planas, Xevi Biarnés, A practical guide to computational tools for engineering biocatalytic properties, Int. J. Mol. Sci. 2025, 26(3), 980
Bastien Bissaro, Julien Durand, Xevi Biarnés, Antoni Planas, Pierre Monsan, Michael J. O’Donohue, Régis Fauré, Molecular Design of Non-Leloir Furanose-Transferring Enzymes from an α-l-Arabinofuranosidase: A Rationale for the Engineering of Evolved Transglycosylases. ACS Catalysis 2015, 5(8), 4598–4611
This research has been conducted within the framework of the GLYCODESIGN (PID2019-104350RB-I00) and GLYCOENGIN (PID2022-138252OB-I00) projects, funded by the Ministry of Science, Innovation and Universities – State Research Agency, within the call for State Programmes for Knowledge Generation and the Scientific and Technological Strengthening of the R&D&i system aimed at the challenges society faces. It has received additional funding from the Obra Social “laCaixa” through the Ramon Llull University research aid programme (Grant: BINDSCAN 2.0’2020).